mlRho version 2.9: Software for Estimating the Population
Mutation and Recombination Rates from Shotgun-Sequenced Diploid Genomes
mlRho
takes as input a file with assembled reads from a single
diploid individual and returns
maximum likelihood estimates of the population mutation rate,
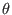 , the sequencing error
, the sequencing error 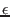 , the zygosity correlation,
, the zygosity correlation,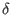 ,
and the population recombination rate,
,
and the population recombination rate, 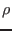 [1]. Please consult the
documentation for further details.
[1]. Please consult the
documentation for further details.
Ciona intestinalis profiles [2].
- 1
-
B. Haubold, P. Pfaffelhuber, and M. Lynch.
mlRho: A program for estimating the population mutation and
recombination rates from shotgun-sequenced diploid genomes.
Molecular Ecology, 19:277-284, 2010.
- 2
-
J.H. Kim, M.S. Waterman, and L.M. Li.
Diploid genome reconstruction of Ciona intestinalis and
comparative analysis with Ciona savignyi.
Genome Res, 17(7):1101-1110, Jul 2007.
mlRho version 2.9: Software for Estimating the Population
Mutation and Recombination Rates from Shotgun-Sequenced Diploid Genomes
This document was generated using the
LaTeX2HTML translator Version 2008 (1.71)
Copyright © 1993, 1994, 1995, 1996,
Nikos Drakos,
Computer Based Learning Unit, University of Leeds.
Copyright © 1997, 1998, 1999,
Ross Moore,
Mathematics Department, Macquarie University, Sydney.
The command line arguments were:
latex2html -split 0 -local_icons mlRho -image_type gif
The translation was initiated by Bernhard Haubold on 2016-09-22
Bernhard Haubold
2016-09-22