Several scripts for visualizing the output of hitchhiking are
included with the source code. These scripts run under the statistics
package R. The example graphs all correspond to figures in the paper for which we
developed hitchhiking []:
- heterozygosity.r: reduction in heterozygosity depending on the
recombination rate and the selection strength (Figure 2);
- time1.r: runtime ration depending on
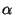 (Figure 3a);
(Figure 3a);
- time2.r: runtime ration depending on n (Figure 3b);
- smallb.r: events in the wild-type background depending on
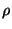 (Figure 4a);
(Figure 4a);
- smallb2.r: events in the wild-type background depending on
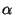 (Figure 4b);
(Figure 4b);
- appErr1.r and appErrEx1.r: approximation error for fixed
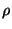 /
/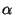 (Figures 5a and 6a);
(Figures 5a and 6a);
- appErr2.r: approximation error for various
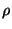 /
/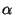 (Figures 5b and 6b);
(Figures 5b and 6b);
- tajima.r: distribution of Tajima's D (Figure 7a);
- likelihood.r: log-likelihood curve for the position of the selected site (Figure 7b).
All of these scripts call routines contained in hitchhiking.r.